

Stéfan van der Walt, University of Stellenbosch
>>> import numpy as np
>>> np.random.seed([1, 2, 3])







Broadcasting
Structured arrays
Views
Sub-classing
(We'll skip this today)
Optimisation tricks
Gems
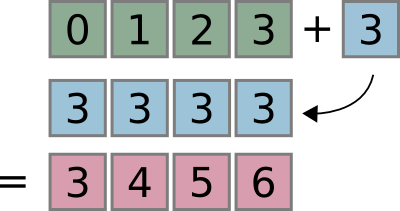
>>> x = np.arange(4)
>>> x
array([0, 1, 2, 3])
>>> x + 3
array([3, 4, 5, 6])
>>> x = np.zeros((3,4))
>>> y = np.zeros((3,1))
>>> (x + y).shape
(3, 4)
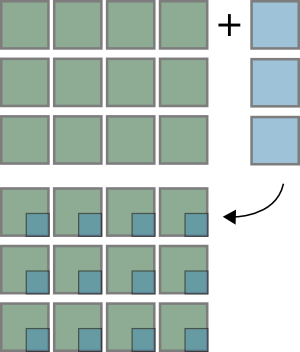
Creating a grid of values:
>>> x, y = np.ogrid[1:10, 1:5]
>>> x
array([[1],
[2],
[3],
[4],
[5],
[6],
[7],
[8],
[9]])
>>> y
array([[1, 2, 3, 4]])
>>> x + 1j*y
array([[ 1.+1.j, 1.+2.j, 1.+3.j, 1.+4.j],
[ 2.+1.j, 2.+2.j, 2.+3.j, 2.+4.j],
[ 3.+1.j, 3.+2.j, 3.+3.j, 3.+4.j],
[ 4.+1.j, 4.+2.j, 4.+3.j, 4.+4.j],
[ 5.+1.j, 5.+2.j, 5.+3.j, 5.+4.j],
[ 6.+1.j, 6.+2.j, 6.+3.j, 6.+4.j],
[ 7.+1.j, 7.+2.j, 7.+3.j, 7.+4.j],
[ 8.+1.j, 8.+2.j, 8.+3.j, 8.+4.j],
[ 9.+1.j, 9.+2.j, 9.+3.j, 9.+4.j]])
Timing two different approaches:
>>> def grid_old_way():
... x, y = np.mgrid[1:100, 1:100]
... return x + 1j*y
>>> def grid_new_way():
... x, y = np.ogrid[1:100, 1:100]
... return x + 1j*y
>>> timeit grid_new_way()
1000 loops, best of 3: 301 µs per loop
>>> timeit grid_old_way()
1000 loops, best of 3: 1.44 ms per loop
>>> x = np.zeros((3, 5))
>>> y = np.zeros(8)
>>> (x[..., None] + y).shape
(3, 5, 8)
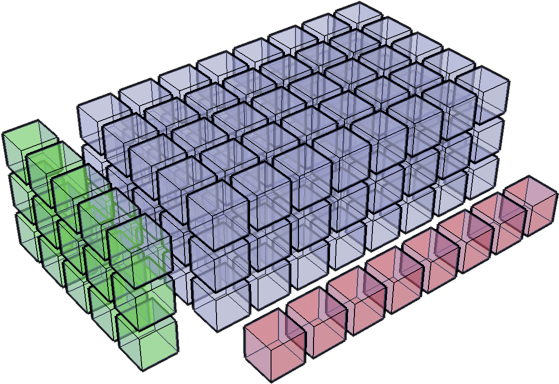
Compare dimensions, starting from last
Match when either dimension is one or None, or if dimensions are equal
Scalar 2D 3D No go ( ,) (3, 4) (3, 5, 1) (3, 5, 2) (3,) (3, 1) ( 8) ( 8) ---- ------ ---------- --------- (3,) (3, 4) (3, 5, 8) XXX
>>> book = np.arange(1000).reshape(100,10)
>>> match = np.array([np.arange(100, 110),
... np.arange(155, 165),
... np.arange(173, 183)])
>>> for v in match:
... min_dist = np.inf
... for d in book:
... dist = np.sum((v - d)**2)
... if dist < min_dist:
... best_match = d
... min_dist = dist
... print best_match, min_dist
[100 101 102 103 104 105 106 107 108 109] 0
[150 151 152 153 154 155 156 157 158 159] 250
[170 171 172 173 174 175 176 177 178 179] 90
>>> distances = (book - match[:, None, :])
>>> distances *= distances # square in-place
>>> distances = distances.sum(axis=2)
>>> chosen = distances.argmin(axis=1)
>>> book[chosen]
array([[100, 101, 102, 103, 104, 105, 106, 107, 108, 109],
[150, 151, 152, 153, 154, 155, 156, 157, 158, 159],
[170, 171, 172, 173, 174, 175, 176, 177, 178, 179]])
>>> distances[range(len(match)), chosen]
array([ 0, 250, 90])
Not ideal -- the temporary array created has a large memory footprint:
( 100, 10) (3, 1, 10) ------------ (3, 100, 10)
Usually, the number of vectors compared would be much greater, which exacerbates the problem.
Date: Wed, 16 Jul 2008 16:45:37 -0500
From: <Jack.Cook@>
To: <numpy-discussion@scipy.org>
Subject: [Numpy-discussion] Numpy Advanced
Indexing Question
Greetings,
I have an I,J,K 3D volume of amplitude values
at regularly sampled time intervals. I have an
I,J 2D slice which contains a time (K) value at
each I, J location. What I would like to do is
extract a subvolume at a constant +/- K window
around the slice. Is there an easy way to do
this using advanced indexing or some other
method? Thanks in advanced for your help.
- Jack
Robert Kern responds:
cube[:, :, K-half_width:K+half_width]
But Jack's problem turned out to be a bit more tricky:
I can understand how this works if K is a constant
time value but in my case K varies at each
location in the two-dimensional slice. In other
words, if I was doing this in a for loop I would
do something like this
for i in range(numI):
for j in range(numJ):
k = slice(i,j)
trace = cube(i,j,k-half_width:k+half_width)
# shove trace in sub volume

First part of Robert's answer, shortened for tutorial purposes:
It would be great if you could just use slices
for the first two axes:
cube[:, :, slice + numpy.arange(-half_width,
half_width)]
but the semantics of that are a bit different
for reasons I can explain later, if you want.
Why doesn't this work?
Diagonal
| 1 | 2 | 3 |
| 4 | 5 | 6 |
| 7 | 8 | 9 |
>>> x = np.arange(9).reshape((3,3))
>>> x
array([[0, 1, 2],
[3, 4, 5],
[6, 7, 8]])
>>> x[:, [0, 1, 2]]
array([[0, 1, 2],
[3, 4, 5],
[6, 7, 8]])
>>> np.array([x[:, 0], x[:, 1], x[:, 2]])
array([[0, 3, 6],
[1, 4, 7],
[2, 5, 8]])
Safety First!
Prepare Your Foil Deflector Beanies
Two-step process to predict the outcome:
>>> x = np.random.random((15,12,16,3))
>>> index_one = np.array([[0, 1], [2, 3], [4, 5]])
>>> index_one.shape
(3, 2)
>>> index_two = np.array([[0, 1]])
>>> index_two.shape
(1, 2)
x[5:10, index_one, :, index_two].shape
>>> x = np.random.random((15,12,16,3))
>>> index_one = np.array([[0, 1], [2, 3], [4, 5]])
>>> index_one.shape
(3, 2)
>>> index_two = np.array([[0, 1]])
>>> index_two.shape
(1, 2)
Broadcast index_one and index_two:
(3, 2) # shape of index_one (1, 2) # shape of index_two ------ (3, 2)
Keeping this in mind, we may just be able to predict the following output:
>>> x[5:10, index_one, :, index_two].shape
(3, 2, 5, 16)
TIP: Use numpy.newaxis to change array dimensions to appease the Big Broadcaster.
>>> ni, nj, nk = (10, 15, 20)
# Make a fake data cube such that cube[i,j,k] == k for all i,j,k.
>>> cube = np.empty((ni,nj,nk), dtype=int)
>>> cube[:,:,:] = np.arange(nk)[np.newaxis, np.newaxis, :]
# Pick out a random fake horizon in k.
>>> kslice = np.random.randint(5, 15, size=(ni, nj))
>>> kslice
array([[ 6, 9, 11, 10, 9, 10, 8, 13, 10, 12, 13, 9, 12, 5, 6],
[ 7, 9, 6, 14, 11, 8, 12, 7, 12, 9, 7, 9, 8, 10, 13],
[10, 14, 9, 13, 12, 11, 13, 6, 11, 9, 14, 12, 6, 8, 12],
[ 5, 11, 8, 14, 10, 10, 10, 9, 10, 5, 7, 11, 9, 13, 8],
[ 7, 8, 8, 5, 13, 9, 11, 13, 13, 12, 13, 11, 12, 5, 11],
[11, 9, 13, 14, 6, 7, 6, 14, 10, 6, 8, 14, 14, 14, 14],
[10, 12, 6, 7, 8, 6, 10, 9, 13, 6, 14, 10, 12, 10, 10],
[10, 12, 10, 9, 11, 14, 9, 6, 7, 13, 6, 11, 8, 11, 8],
[13, 14, 7, 14, 6, 14, 6, 8, 14, 7, 14, 12, 8, 5, 10],
[13, 5, 9, 7, 5, 9, 13, 10, 13, 7, 7, 9, 14, 13, 11]])
>>> half_width = 3
# These two replace the empty slices for the first two axes.
>>> idx_i = np.arange(ni)[:, np.newaxis, np.newaxis]
>>> idx_j = np.arange(nj)[np.newaxis, :, np.newaxis]
# This is the substantive part that picks out the window
>>> idx_k = kslice[:, :, np.newaxis] + \
... np.arange(-half_width, half_width+1) # (10, 15, 7)
cube[idx_i, idx_j, idx_k]
Apply the rule -- broadcast array indices, then add slices (but there are no slices in this case):
(ni, 1, 1 ) # idx_i (1 , nj, 1 ) # idx_j (ni, nj, 2*half_width + 1 ) # idx_k --------------------------- (ni, nj, 7)
Phew, tough one. Moving on to easier things...
>>> smallcube = cube[idx_i,idx_j,idx_k]
>>> smallcube.shape
(10, 15, 7)
# Now verify that our window is centered on kslice everywhere:
>>> smallcube[:,:,3]
array([[ 6, 9, 11, 10, 9, 10, 8, 13, 10, 12, 13, 9, 12, 5, 6],
[ 7, 9, 6, 14, 11, 8, 12, 7, 12, 9, 7, 9, 8, 10, 13],
[10, 14, 9, 13, 12, 11, 13, 6, 11, 9, 14, 12, 6, 8, 12],
[ 5, 11, 8, 14, 10, 10, 10, 9, 10, 5, 7, 11, 9, 13, 8],
[ 7, 8, 8, 5, 13, 9, 11, 13, 13, 12, 13, 11, 12, 5, 11],
[11, 9, 13, 14, 6, 7, 6, 14, 10, 6, 8, 14, 14, 14, 14],
[10, 12, 6, 7, 8, 6, 10, 9, 13, 6, 14, 10, 12, 10, 10],
[10, 12, 10, 9, 11, 14, 9, 6, 7, 13, 6, 11, 8, 11, 8],
[13, 14, 7, 14, 6, 14, 6, 8, 14, 7, 14, 12, 8, 5, 10],
[13, 5, 9, 7, 5, 9, 13, 10, 13, 7, 7, 9, 14, 13, 11]])
>>> (smallcube[:,:,3] == kslice).all()
True
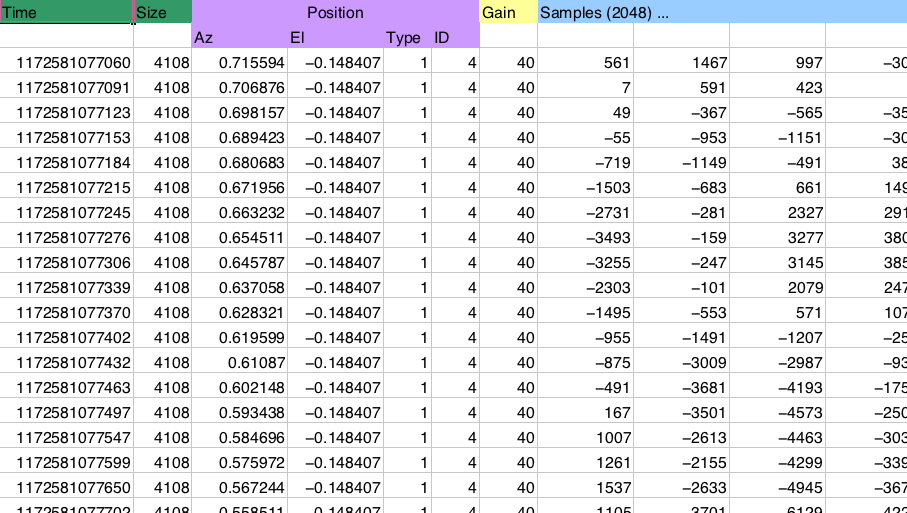
Reading this in most languages requires somewhat messy code. Not too bad, but not very pretty either:
while ((count > 0) && (n <= NumPoints))
% get time - I8 [ms]
[lw, count] = fread(fid, 1, 'uint32');
if (count > 0) % then carry on
uw = fread(fid, 1, 'int32');
t(1,n) = (lw+uw*2^32)/1000;
% get number of bytes of data
numbytes = fread(fid, 1, 'uint32');
% read sMEASUREMENTPOSITIONINFO (11 bytes)
m(1,n) = fread(fid, 1, 'float32'); % az [rad]
m(2,n) = fread(fid, 1, 'float32'); % el [rad]
m(3,n) = fread(fid, 1, 'uint8'); % region type
m(4,n) = fread(fid, 1, 'uint16'); % region ID
g(1,n) = fread(fid, 1, 'uint8');
numsamples = (numbytes-12)/2; % 2 byte integers
a(:,n) = fread(fid, numsamples, 'int16');
The NumPy solution:
dt = np.dtype([('time', np.uint64),
('size', np.uint32),
('position', [('az', np.float32),
('el', np.float32),
('region_type', np.uint8),
('region_ID', np.uint16)]),
('gain', np.uint8),
('samples', (np.int16,2048))])
data = np.fromfile(f, dtype=dt)
Access this data using dictionary-like syntax:
data['position']['az']
Date: Wed, 16 Jul 2008 16:45:37 -0500
From: Amir
To: <numpy-discussion@scipy.org>
Subject: [Numpy-discussion] interleaved
indexing
A very beginner question about indexing: let x be
an array where n = len(x). I would like to create
a view y of x such that:
y[i] = x[i:i+m,...] for each i and a fixed m << n
so I can do things like numpy.cov(y). With n
large, allocating y is a problem for
me. Currently, I either do for loops in cython or
translate operations into correlate() but am
hoping there is an easier way, maybe using fancy
indexing or broadcasting. Memory usage is
secondary to speed, though.
Given input data:
>>> x = np.arange(20).reshape([4, 5])
>>> x
array([[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19]])
What he wants is a view like this (assume m == 2):
array([[[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9]],
[[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14]],
[[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19]]])
To do this, we need to know the following terms:
>>> x.strides
(20, 4)
>>> np.int32().itemsize
4
array([[[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9]],
[[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14]],
[[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19]]])
We now need to manipulate the array shape and strides. The output shape must be (3, 2, 5), i.e. 3 items, each containing two rows (m == 2), and each row having 5 elements.
The strides need to change from (20, 4), to (20, 20, 4). Each item in the new output array starts at a new row, that each row consists of 20 bytes (5 elements of 4 bytes each), and each element occupies 4 bytes (int32).
Let's do it:
>>> from numpy.lib import stride_tricks
>>> stride_tricks.as_strided(x, shape=(3, 2, 5),
... strides=(20, 20, 4))
array([[[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9]],
[[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14]],
[[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19]]])
Or the old way (which is basically what as_strided does under the hood):
>>> d = dict(x.__array_interface__)
>>> d['shape'] = (3, 2, 5)
>>> d['strides'] = (20, 20, 4)
>>> class Arr:
... __array_interface__ = d
... base = x
>>> np.array(Arr())
array([[[ 0, 1, 2, 3, 4],
[ 5, 6, 7, 8, 9]],
[[ 5, 6, 7, 8, 9],
[10, 11, 12, 13, 14]],
[[10, 11, 12, 13, 14],
[15, 16, 17, 18, 19]]])
Two important methods:
class InfoArray(np.ndarray):
def __new__(subtype, data, info=None, dtype=None, copy=False):
subarr = np.array(data, dtype=dtype, copy=copy)
subarr = subarr.view(subtype)
if info is not None:
subarr.info = info
elif hasattr(data, 'info'):
subarr.info = data.info
return subarr
def __array_finalize__(self, obj):
self.info = getattr(obj, 'info', {})
I would like to thank
